Written by
C
ancer is the term used to describe a group of complex genetic diseases that result in the uncontrolled division of abnormal cells. Cancer cells can arise from specific genetic mutations in a single cell or group of cells. These cells have the potential to invade nearby tissues and, in some cases, metastasize to distant sites within the body via the circulatory or lymphatic system, giving rise to secondary tumors.
Tumors can have distinct underlying genetic origins and may express different proteins in one patient versus another. Analyzing a patient’s omics data – including genomics, proteomics, metabolomics and transcriptomics – can help to determine their risk of developing cancer, enable early detection of the disease and identify targeted drugs or drug combinations with optimal efficacy and minimal adverse effects. This individualized approach is known as personalized medicine.
T
hanks to advances in genomics technologies such as DNA sequencing, it is now possible to rapidly sequence an individual’s whole genome or deeply sequence specific target regions of a genome to identify genetic variants that contribute to disease risk, or can help to diagnose, treat and better understand a patient’s cancer. While genomics techniques have been at the forefront of personalized medicine for some time, additional omics approaches are now more widely adopted.
RNA sequencing (RNAseq) is being harnessed to study the transcriptional landscape of human cancers. Whole transcriptome RNAseq can help to guide treatment decisions and identify biomarkers for cancer progression. Proteomics techniques, such as mass spectrometry, are enabling researchers to analyze the complexity of cellular physiology, including protein expression patterns, the presence of isoforms, post-translational modifications and protein–protein interactions – which can all impact an individual’s response to a disease or treatment as many drugs targets are proteins. Metabolomics approaches can help to develop cancer diagnostic methods centered on the characterization of metabolic subtypes. Metabolomics can also be used to investigate an individual’s response to a drug or to identify key mechanisms involved in disease relapse or drug resistance. Mass-spectrometry imaging is being used to understand the tumor landscape at a spatial level, helping to identify tumor-associated metabolite and enzyme alterations.
Different types of patient-derived cancer models have been developed to help predict a patient’s response to a drug. One of the most advanced in vitro models is the patient-derived organoid (PDO). PDOs are three-dimensional cell cultures that resemble many aspects of the original tumor. Due to their ability to imitate pathogenic tissue, drug responses observed in patient-derived cancer organoids often mirror patients’ clinical response.
A tumor biopsy is obtained from a cancer patient and a cancer model is derived from the tumor cells. In parallel, sequencing of the tumor is performed, and the data is analyzed to identify potential gene–drug associations. High-throughput drug screening is conducted and compared to the gene–drug association data to determine whether a patient has specific drug sensitivities.
W
hile personalized medicine is a rapidly evolving field, several challenges need to be addressed before it can be implemented at a large scale in the oncology field.
1. Data overload. The emergence of data-intensive omics and imaging techniques has led to the generation of huge amounts of data. Processing and storing this data is a key bottleneck and consequently there is an urgent need to develop scalable solutions that can enable the rapid deposit, retrieval and analysis of data. In addition, to avoid data silos, and to enable effective analysis of all available information, data quality is data standardization is crucial.
2. Time and cost. Identifying and/or designing therapeutics that can target the drivers of specific cancers takes time and a considerable amount of money. The number of patients with a cancer type containing a specific “driver” can be extremely low, meaning it may not be financially viable for companies to develop therapeutics that target it. Getting a drug from bench to bedside can take 10–15 years, therefore even in instances where a cancer driver is known and many patients are affected, the targeted therapy may not become clinically available for some time.
3. Drug resistance. Some targeted drugs, while initially successful, do not provide long-term benefit due to the patient developing resistance to the therapy. Cancer cells are extremely adaptable and can induce mechanisms that induce direct or indirect drug resistance. Examples include; drug inactivation, alteration of the drug target, inhibition of cell death and the repair of DNA damage.
4. Medical ethics and informed consent. As more personalized medicines move towards clinical development and approval, attention needs to be focused towards related ethical, legal and social implications. How will we cope with the increase in health information brought about by personalized medicine? Could personalized medicine intensify existing disparities in healthcare? How do we ensure privacy and confidentiality? How will informed consent be handled for patients undergoing genetic testing?
T
argeted cancer drugs “target” the genetic differences between cancer cells and normal cells. Two patients diagnosed with the same cancer may have genetic variants that could influence their individual response to these drugs – a field of research known as pharmacogenomics. Genetic testing of tumor tissue and/or liquid biopsies can help to identify specific targets, helping to “match” a patient to a personalized treatment strategy. Here, we list some key examples of these therapies along with their targets.
Find out about particular cancers by clicking the highlighted areas on the image below.

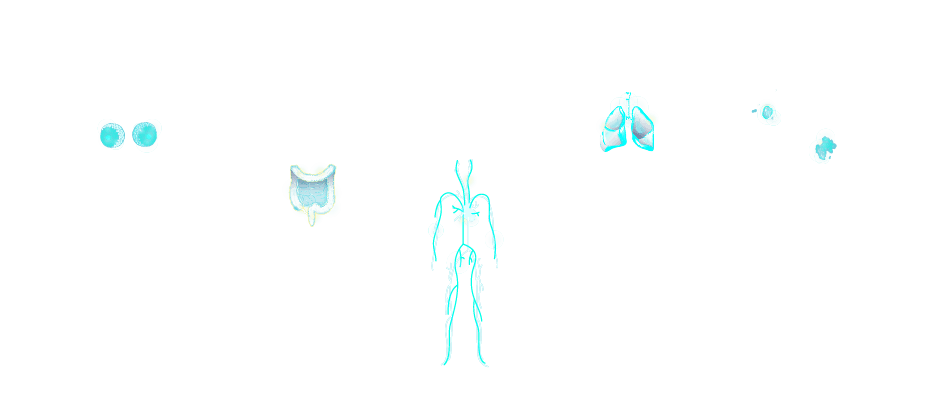
Breast Cancer. A mutation in the HER2 gene results in the overexpression of the human epidermal growth factor receptor 2 (HER2) in approximately 15–30% of invasive breast cancers. Several targeted treatments are available for HER2-postitive breast cancer.
Colorectal Cancer. Approximately 1 in 10 colorectal cancer patients has a mutation in BRAF. As the median survival for these patients is less than one year, identifying individuals in this subgroup is key to developing tailored treatment strategies beyond standard therapy.
Childhood Leukemia. Mercaptopurine has been shown to be effective at treating childhood acute lymphoblastic leukemia. However, the toxicity of the drug is variable due to genetic mutations in genes coding for enzymes responsible for metabolizing mercaptopurine. One such gene is TPMT.
Lung Cancer. Approximately 5% of non-small cell lung cancers possess a rearrangement in ALK. The resulting abnormal ALK protein can be successfully targeted with a variety of drugs.
Melanoma. Approximately 50% of melanomas have a BRAF mutation. Patients confirmed as having BRAF V600-mutated melanoma have been shown to benefit from selective inhibition of the MAPK signaling using BRAF and MEK inhibitors.
1. Breast Cancer. A mutation in the HER2 gene results in the overexpression of the human epidermal growth factor receptor 2 (HER2) in approximately 15–30% of invasive breast cancers. Breast cancer cells can have a 40–100-fold increase in HER2 protein, leading to the expression of ~ 2 million receptors on the cell surface. For individuals diagnosed with HER2-postitive breast cancer, several targeted treatments are available, including trastuzumab, margetuximab and pertuzumab.
2. Colorectal Cancer. Epidermal growth factor receptor (EGFR) is often overexpressed in colorectal cancers, meaning drugs targeting EGFR, such as cetuximab and panitumumab, could be effective treatment strategies. However, in patients with a mutated KRAS gene, these two drugs are ineffective. Therefore, to determine the suitability of the treatment, genetic testing is often carried out first to confirm if a patient with colorectal cancer has a mutation in KRAS. Approximately one in ten patients with colorectal cancer has a mutation in BRAF. As the median survival for these patients is less than one year, identifying individuals in this subgroup is key to developing tailored treatment strategies beyond anti-EGFR therapy. The BRAF inhibitor encorafenib in combination with cetuximab has been shown to improve survival of colorectal cancer patients with mutated BRAF.
3. Leukemia. The chemotherapy drug mercaptopurine has been shown to be highly effective at treating childhood acute lymphoblastic leukemia (ALL). However, the toxicity of the drug is variable due to genetic polymorphisms in genes coding for enzymes responsible for metabolizing mercaptopurine. One such gene is TPMT. Genetic testing of pediatric ALL patients for a genetic mutation in TPMT enables dosing recommendations to be developed to optimize and personalize prescribing of the drug and reduce adverse effects. A translocation in chromosome 22 is responsible for > 90% of chronic myeloid leukemia (CML) cases. This translocation causes the fusion of two genes – BCR and ABL1 – and results in the development of CML. Imatinib, a selective inhibitor, was designed to treat CML by specifically targeting BCR-ABL1 tyrosine kinase, which arises from the chimeric fusion gene BCR-ABL.
4. Lung Cancer. Some patients with lung adenocarcinoma possess activating mutations in the EGFR gene. Patients with mutated EGFR have been successfully treated with several selective tyrosine kinase inhibitors including erlotinib and gefitinib. Approximately 5% of non-small cell lung cancers (NSCLCs) possess a rearrangement in the ALK gene. The resulting abnormal ALK protein can be targeted with a variety of drugs including crizotinib, ceritinib, alectinib, brigatinib and lorlatinib. Targeted therapies have also been developed for NSCLCs with mutations in the following genes: ROS1, RET, MET, NTRK and BRAF.
5. Melanoma. Inhibition of Raf/MEK/ERK and PI3K/AKT/mTOR signaling elements has been identified as a key treatment strategy for melanoma. Studies have shown that aberrant PI3K signaling occurs in a high percentage of melanomas, and the AKT gene is amplified in ~ 45% . Approximately 50% of melanomas have a BRAF mutation – V600E being the most common. Mutated BRAF results in an abnormal version of the BRAF kinase. Patients confirmed as having BRAF V600-mutated melanoma have been shown to benefit from selective inhibition of the MAPK signaling using BRAF and MEK inhibitors. Patients can acquire resistance to single-agent BRAF inhibitors. However, BRAF and MEK inhibitor combination therapy has been shown to improve treatment response and delay resistance.
While personalized medicine is driving a dramatic revolution in clinical practice, attention must be given to the numerous challenges remain to be fully addressed.
Only then can the true potential of personalized medicine be realized – and its broader application be achieved.